I’ve spent a bit of time trying to learn how to run a program called BUGS. The acronym stands for Bayes Using Gibbs Sampling. Here is my first serious attempt to run a BUGS program.
There are several web pages about this program, most notably The BUGS Project Welcome Page. That page had links to several resources about BUGS, including a recently published book by George G. Woodworth, Biostatistics: A Bayesian Introduction. The author placed a PDF version of the Appendix describing how to use WinBUGS, the Windows version of BUGS on the web.
There is also a version of BUGS that works under R, called BRugs. A nice description of BRugs appears at the website devoted to the book by Andrew Gelman, John B. Carlin, Hal S. Stern, and Donald B. Rubin, Bayesian Data Analysis." In particular, the page offers some very useful advice.
Dr. Woodworth starts with the simplest of possible examples, the beta-binomial model. To fit this, you need to prepare a model statement
MODEL {
p ~ dbeta(a,b)
x ~ dbin(p,n)
}
This model specifies that the number of successes x is represented by a binomial distribution with probability of success p and number of trials n. It also specifies a prior distribution for p which is beta with parameters alpha and beta equal to a and b, respectively.
You also need a data statement
DATA list(a=1,b=1,x=11,n=25)
This data list provides information about the observed data. You collected data on 25 subjects and 11 of them were classified as success. It also provides specifications for the prior distribution. The values a=1 and b=1 produce a beta(1,1) distribution which is also known as a uniform distribution. This represents a “flat” or uninformative prior. Data analysts select a flat prior when they have little information prior to the start of the study about the true value of p or if they prefer to let the observed data provide most of the weight in the resulting posterior distribution.
The steps in running BUGS are
- check your syntax,
- load the data set,
- compile your model,
- generate initial values,
- monitor the nodes,
- update the model,
- exclude the early samples,
- calculate summary statistics.
Here are the eight steps in a BUGS model using the beta-binomial model.
Step 1. check your syntax.
In WinBUGS, you need to open the Specification Tool dialog box
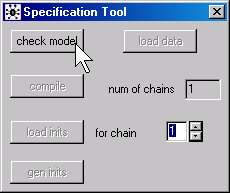
by selecting Model | Specification from the menu. Highlight any set of characters inside the curly braces of the MODEL statement and then click on the check model button. In BRugs, you need to store the model as a text file and then call the function:
checkModel("BBModel.txt")
In either case, you should get a message that reads
model is syntactically correct
Step 2. load the data set.
In WinBUGS, highlight the word “list” and click on the load data button. In BRugs, you need to store the data as a text file and then call the function
modelData("BBData.txt")
In either case, you should get a message that reads
data loaded
Step 3. compile your model.
In WinBUGS, click on the compile button. In BRugs, call the function
modelCompile()
You will see a message
model compiled
Step 4. Generate initial values.
The simulation methods in BUGS require starting values. For simple models, BUGS can generate those starting values for you, but for more complex models, you have to provide starting values. The beta-binomial model is simple enough that BUGS can do this.
In WinBUGs, click on the gen inits button and in BRugs, call the function
modelGenInits()
You will see a message
initial values generated, model initialized
Step 5. monitor the nodes.
In WinBUGS, open the Sample Monitor Tool dialog box
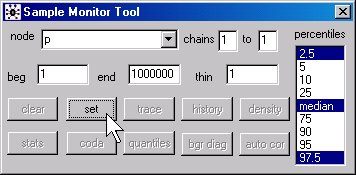
by selecting Inference | Samples from the menu. Type “p” in the node field and click on the set button. In BRugs, call the function
samplesSet("p")
You will get the diagnostic message
monitor set for variable 'p'
Step 6. update the model.
In WinBUGS, open the Update Tool dialog box
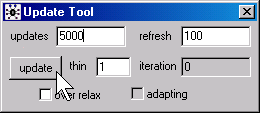
by selecting Model | Update from the menu. For this analysis, 5,000 is a reasonable value for the updates field. Click on the update button after you have made this change. In BRugs, call the function
modelUpdate(5000)
You will see the diagnostic message
5000 updates took 0 s
That was fast!
Step 7. exclude the early values.
The Markov Chain is dependent on the initial starting value and sometimes takes a while to settle down to a process that reflects the true probability distribution (this is called a steady state). The early values in the Markov Chain may thus exhibit some artefactual patterns that can provide misleading results. You will often get better simulation results if you ignore the first portion of the chain. In WinBUGS, adjust the Sample Monitor Tool dialog box to exclude the first thousand samples by changing the beg field from 1 to 1000. In BRugs, call the function
samplesSetBeg(1000)
Step 8. calculate summary statistics.
In WinBUGS, click on the stats button in the Sample Monitor Tool dialog box. In BRugs, call the function
samplesStats("p")
This produces the output
mean sd MC_error val2.5pc median val97.5pc start sample
p 0.4421 0.09435 0.001237 0.2634 0.441 0.6286 1000 5000
What do these numbers mean?
In this example, you observed x=11 successes in a sample of size n=25. The classical estimate of the probability of success would be 11/25=0.44. In the Bayesian example, your prior distribution is beta(1,1). A simple algebraic solution for the posterior distribution is easy enough to find (it is a beta distribution with alpha=12 and beta=15), but using WinBUGS or BRugs, you can also illustrate how a simulation could produce roughly the same answer.
The value labeled “mean” represents the mean of the posterior distribution. The simulation estimates this value to be 0.4421. If you used the algebraic solution, you would get a posterior mean of 0.4444. The value labeled “sd” represents the standard deviation of the posterior distribution.
The value labeled “MC_error” is the uncertainty associated with this particular simulation. If this value is much smaller than the standard deviation of the posterior distribution, then your simulation has done well. If it is not at least an order of magnitude smaller, then you should consider changing the simulation (e.g., increasing the number of samples).
The values labeled “val2.5pc” and “val97.5pc” represent an interval that contains 95% of the posterior distribution. This interval is called the Credible Interval (CrI) and represents the Bayesian alternative to a confidence interval. The simulated values 0.2634 and 0.6286 show that there is a fair amount of uncertainty about the true proportion p, which is hardly surprising given the small sample size. These values for the CrI computed by the simulation compare nicely with the exact values from the algebraic solution (0.2659 and 0.6308).
The value labeled “median” represents the median of the posterior distribution. In this example, the posterior distribution is almost symmetric, so this value differs only slightly from the mean of the posterior distribution. The simulated value of the median (0.4410) and the exact value from the algebraic solution (0.4431) are very close.
The values labeled “start” and “sample” simply remind you what you set for the beginning and end of the Markvo chain simulation
Combining these steps
In BRugs, you can combine these steps by using the function BRugsFit. Here’s an example:
BBOutput <- BRugsFit(modelFile="BBModel.txt",data="BBData.txt",
numChains=1,parametersToSave="p",nBurnin=1000,nIter=5000)
The diagnostic messages produced by this function are shown below:
model is syntactically correct
data loaded
model compiled
initial values generated, model initialized
1000 updates took 0 s
deviance set
monitor set for variable 'p'
4000 updates took 0 s
The output from this function is stored in BBOutput. When you list BBOutput, you get the following information:
> BBOutput
$Stats
mean sd MC_error val2.5pc median val97.5pc start sample
p 0.4428 0.09491 0.001376 0.2626 0.441 0.6296 1001 5000
$DIC
Dbar Dhat DIC pD
x 4.641 3.677 5.605 0.9639
total 4.641 3.677 5.605 0.9639`
Notice that the output here is slightly different than the output shown above. The mean, for example, is 0.4428 rather than 0.4421. This should not be too surprising. The BUGS software computes estimates using a simulation, and these change each time you run the program. The difference between the two estimates is about the same order of magnitude as the Monte Carlo error.
Also notice that the BRgusFit function, by default, includes an additional set of statistics known as the Deviance Information Criterion. This statistic is useful for comparing models of differing complexity. There is no way to interpret this value for a single model in isolation.
Further steps.
There are additional graphical and statistical summaries available in both WinBUGS and BRugs. There are also diagnostic tools to help you determine whether the markov chain has settled down sufficiently to provide good estimates.
You can find an earlier version of this page on my old website.