I wanted to show a couple of Cox proportional hazards regression models in R for a talk I am giving for the R users group.
This builds on the work in the previous two blog entries.
This is the documentation of the data file.
# Documentation found at http://lib.stat.cmu.edu/datasets/pbc
# The data set is a study of primary biliary cirrhosis
# Variables:
# V1 case number
# V2 number of days between registration and the earlier of death,
# transplantion
- or study analysis time in July
- 1986
# V3 status
# 0=censored
- 1=censored due to liver tx
- 2=death
# V4 drug
# 1= D-penicillamine
- 2=placebo
# V5 age in days
# V6 sex
# 0=male
- 1=female
# V7 presence of asictes
# 0=no 1=yes
# V8 presence of hepatomegaly
# 0=no 1=yes
# V9 presence of spiders
# 0=no 1=yes
# V10 presence of edema
# 0 = no edema and no diuretic therapy for edema;
# 0.5 = edema present without diuretics
- or edema resolved by diuretics;
# 1 = edema despite diuretic therapy
# V11 serum bilirubin in mg/dl
# V12 serum cholesterol in mg/dl
# V13 albumin in gm/dl
# V14 urine copper in ug/day#
# V15 alkaline phosphatase in U/liter
# V16 SGOT in U/ml
# V17 triglicerides in mg/dl
# V18 platelets per cubic ml / 1000
# V19 prothrombin time in seconds
# V20 histologic stage of disease
The dataset is already part of the survival package.
library("survival")
## Loading required package: splines
head(pbc)
## id time status trt age sex ascites hepato spiders edema bili chol
## 1 1 400 2 1 58.76523 f 1 1 1 1.0 14.5 261
## 2 2 4500 0 1 56.44627 f 0 1 1 0.0 1.1 302
## 3 3 1012 2 1 70.07255 m 0 0 0 0.5 1.4 176
## 4 4 1925 2 1 54.74059 f 0 1 1 0.5 1.8 244
## 5 5 1504 1 2 38.10541 f 0 1 1 0.0 3.4 279
## 6 6 2503 2 2 66.25873 f 0 1 0 0.0 0.8 248
## albumin copper alk.phos ast trig platelet protime stage
## 1 2.60 156 1718.0 137.95 172 190 12.2 4
## 2 4.14 54 7394.8 113.52 88 221 10.6 3
## 3 3.48 210 516.0 96.10 55 151 12.0 4
## 4 2.54 64 6121.8 60.63 92 183 10.3 4
## 5 3.53 143 671.0 113.15 72 136 10.9 3
## 6 3.98 50 944.0 93.00 63 NA 11.0 3
tail(pbc)
## id time status trt age sex ascites hepato spiders edema bili
## 413 413 989 0 NA 35.00068 f NA NA NA 0 0.7
## 414 414 681 2 NA 67.00068 f NA NA NA 0 1.2
## 415 415 1103 0 NA 39.00068 f NA NA NA 0 0.9
## 416 416 1055 0 NA 56.99932 f NA NA NA 0 1.6
## 417 417 691 0 NA 58.00137 f NA NA NA 0 0.8
## 418 418 976 0 NA 52.99932 f NA NA NA 0 0.7
## chol albumin copper alk.phos ast trig platelet protime stage
## 413 NA 3.23 NA NA NA NA 312 10.8 3
## 414 NA 2.96 NA NA NA NA 174 10.9 3
## 415 NA 3.83 NA NA NA NA 180 11.2 4
## 416 NA 3.42 NA NA NA NA 143 9.9 3
## 417 NA 3.75 NA NA NA NA 269 10.4 3
## 418 NA 3.29 NA NA NA NA 350 10.6 4
pmod <- pbc[1:312,]
tail(pmod)
## id time status trt age sex ascites hepato spiders edema bili
## 307 307 1149 0 2 30.57358 f 0 0 0 0 0.8
## 308 308 1153 0 1 61.18275 f 0 1 0 0 0.4
## 309 309 994 0 2 58.29979 f 0 0 0 0 0.4
## 310 310 939 0 1 62.33265 f 0 0 0 0 1.7
## 311 311 839 0 1 37.99863 f 0 0 0 0 2.0
## 312 312 788 0 2 33.15264 f 0 0 1 0 6.4
## chol albumin copper alk.phos ast trig platelet protime stage
## 307 273 3.56 52 1282 130 59 344 10.5 2
## 308 246 3.58 24 797 91 113 288 10.4 2
## 309 260 2.75 41 1166 70 82 231 10.8 2
## 310 434 3.35 39 1713 171 100 234 10.2 2
## 311 247 3.16 69 1050 117 88 335 10.5 2
## 312 576 3.79 186 2115 136 149 200 10.8 2
summary(pmodtime)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 41 1191 1840 2006 2697 4556
table(pmodstatus)
##
## 0 1 2
## 168 19 125
p.surv <- Surv(pmodtime,pmod$status==2)
print(p.surv)
## [1] 400 4500+ 1012 1925 1504+ 2503 1832+ 2466 2400 51 3762
## [12] 304 3577+ 1217 3584 3672+ 769 131 4232+ 1356 3445+ 673
## [23] 264 4079 4127+ 1444 77 549 4509+ 321 3839 4523+ 3170
## [34] 3933+ 2847 3611+ 223 3244 2297 4467+ 1350 4453+ 4556+ 3428
## [45] 4025+ 2256 2576+ 4427+ 708 2598 3853 2386 1000 1434 1360
## [56] 1847 3282 4459+ 2224 4365+ 4256+ 3090 859 1487 3992+ 4191
## [67] 2769 4039+ 1170 3458+ 4196+ 4184+ 4190+ 1827 1191 71 326
## [78] 1690 3707+ 890 2540 3574 4050+ 4032+ 3358 1657 198 2452+
## [89] 1741 2689 460 388 3913+ 750 130 3850+ 611 3823+ 3820+
## [100] 552 3581+ 3099+ 110 3086 3092+ 3222 3388+ 2583 2504+ 2105
## [111] 2350+ 3445 980 3395 3422+ 3336+ 1083 2288 515 2033+ 191
## [122] 3297+ 971 3069+ 2468+ 824 3255+ 1037 3239+ 1413 850 2944+
## [133] 2796 3149+ 3150+ 3098+ 2990+ 1297 2106+ 3059+ 3050+ 2419 786
## [144] 943 2976+ 2615+ 2995+ 1427 762 2891+ 2870+ 1152 2863+ 140
## [155] 2666+ 853 2835+ 2475+ 1536 2772+ 2797+ 186 2055 264 1077
## [166] 2721+ 1682 2713+ 1212 2692+ 2574+ 2301+ 2657+ 2644+ 2624+ 1492
## [177] 2609+ 2580+ 2573+ 2563+ 2556+ 2555+ 2241+ 974 2527+ 1576 733
## [188] 2332+ 2456+ 2504+ 216 2443+ 797 2449+ 2330+ 2363+ 2365+ 2357+
## [199] 1592+ 2318+ 2294+ 2272+ 2221+ 2090 2081 2255+ 2171+ 904 2216+
## [210] 2224+ 2195+ 2176+ 2178+ 1786 1080 2168+ 790 2170+ 2157+ 1235
## [221] 2050+ 597 334 1945+ 2022+ 1978+ 999 1967+ 348 1979+ 1165
## [232] 1951+ 1932+ 1776+ 1882+ 1908+ 1882+ 1874+ 694 1831+ 837+ 1810+
## [243] 930 1690 1790+ 1435+ 732+ 1785+ 1783+ 1769+ 1457+ 1770+ 1765+
## [254] 737+ 1735+ 1701+ 1614+ 1702+ 1615+ 1656+ 1677+ 1666+ 1301+ 1542+
## [265] 1084+ 1614+ 179 1191 1363+ 1568+ 1569+ 1525+ 1558+ 1447+ 1349+
## [276] 1481+ 1434+ 1420+ 1433+ 1412+ 41 1455+ 1030+ 1418+ 1401+ 1408+
## [287] 1234+ 1067+ 799 1363+ 901+ 1329+ 1320+ 1302+ 877+ 1321+ 533+
## [298] 1300+ 1293+ 207 1295+ 1271+ 1250+ 1230+ 1216+ 1216+ 1149+ 1153+
## [309] 994+ 939+ 839+ 788+
Run a simple Cox proportional hazards model
cox.mod1 <- coxph(p.surv~sex,data=pmod,na.action=na.omit)
print(cox.mod1)
## Call:
## coxph(formula = p.surv ~ sex
- data = pmod
- na.action = na.omit)
coef exp(coef) se(coef) z p
sexf -0.484 0.616 0.236 -2.05 0.041
Likelihood ratio test=3.77 on 1 df
- p=0.0521 n= 312
- number of events= 125
summary(cox.mod1)
## Call:
## coxph(formula = p.surv ~ sex
- data = pmod
- na.action = na.omit)
n= 312
- number of events= 125
coef exp(coef) se(coef) z Pr(>|z|)
sexf -0.4839 0.6164 0.2365 -2.046 0.0407 *
—
Signif. codes: 0 ‘’ 0.001 ‘’ 0.01 ‘’ 0.05 ‘.’ 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
sexf 0.6164 1.622 0.3877 0.9798
Concordance= 0.532 (se = 0.015 )
Rsquare= 0.012 (max possible= 0.983 )
Likelihood ratio test= 3.77 on 1 df
- p=0.05214
Wald test = 4.19 on 1 df
- p=0.04075
Score (logrank) test = 4.27 on 1 df
- p=0.03887
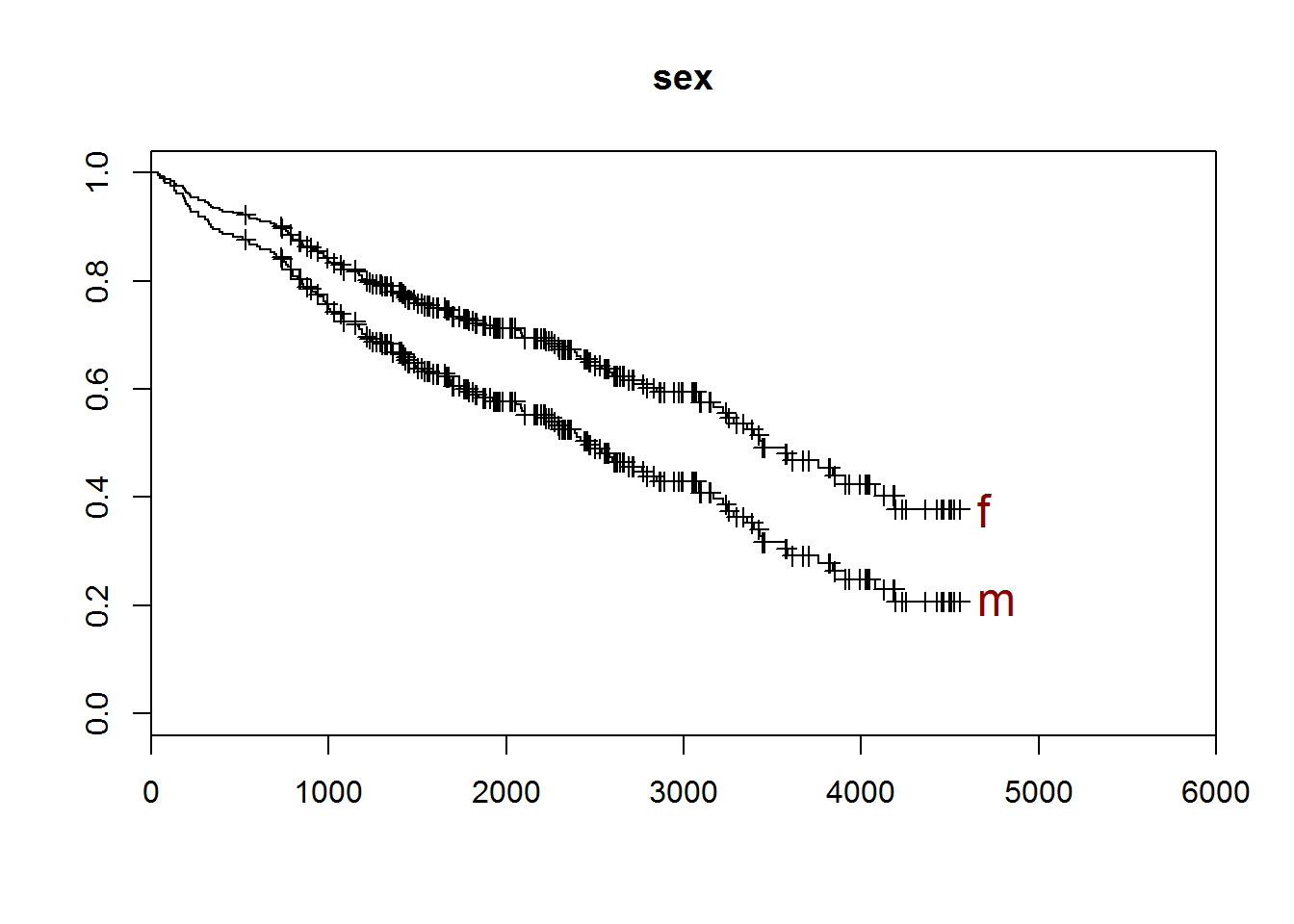
To plot the data
- you need to create predictions at the specific values of interest. With multiple indpendent variables
- any variable left out would be predicted at its average value.
pred.mod1 <- survfit(cox.mod1,newdata=data.frame(sex=c("f","m")))
plot(pred.mod1,xlim=c(0,6000))
title("sex")
end.pts <- dim(pred.mod1surv)[1]
end.x <- rep(pred.mod1time[end.pts]+100,1)
end.y <- pred.mod1surv[end.pts,]
end.nm <- c("f","m")
text(end.x,end.y,end.nm,cex=1.5,col="darkred",adj=0)
The Cox model works fairly similarly for continuous independent variables.
cox.mod1 <- coxph(p.surv~sex,data=pmod,na.action=na.omit)
print(cox.mod1)
## Call:
## coxph(formula = p.surv ~ sex
- data = pmod
- na.action = na.omit)
coef exp(coef) se(coef) z p
sexf -0.484 0.616 0.236 -2.05 0.041
Likelihood ratio test=3.77 on 1 df
- p=0.0521 n= 312
- number of events= 125
summary(cox.mod1)
## Call:
## coxph(formula = p.surv ~ sex
- data = pmod
- na.action = na.omit)
n= 312
- number of events= 125
coef exp(coef) se(coef) z Pr(>|z|)
sexf -0.4839 0.6164 0.2365 -2.046 0.0407 *
—
Signif. codes: 0 ‘’ 0.001 ‘’ 0.01 ‘’ 0.05 ‘.’ 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
sexf 0.6164 1.622 0.3877 0.9798
Concordance= 0.532 (se = 0.015 )
Rsquare= 0.012 (max possible= 0.983 )
Likelihood ratio test= 3.77 on 1 df
- p=0.05214
Wald test = 4.19 on 1 df
- p=0.04075
Score (logrank) test = 4.27 on 1 df
- p=0.03887
cox.mod2 <- coxph(p.surv~age,data=pmod,na.action=na.omit)
print(cox.mod2)
## Call:
## coxph(formula = p.surv ~ age
- data = pmod
- na.action = na.omit)
coef exp(coef) se(coef) z p
age 0.04 1.04 0.00881 4.54 5.7e-06
Likelihood ratio test=20.5 on 1 df
- p=5.95e-06 n= 312
- number of events= 125
summary(cox.mod2)
## Call:
## coxph(formula = p.surv ~ age
- data = pmod
- na.action = na.omit)
n= 312
- number of events= 125
coef exp(coef) se(coef) z Pr(>|z|)
age 0.039995 1.040806 0.008811 4.539 5.65e-06 ***
—
Signif. codes: 0 ‘’ 0.001 ‘’ 0.01 ‘’ 0.05 ‘.’ 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
age 1.041 0.9608 1.023 1.059
Concordance= 0.625 (se = 0.029 )
Rsquare= 0.064 (max possible= 0.983 )
Likelihood ratio test= 20.51 on 1 df
- p=5.947e-06
Wald test = 20.6 on 1 df
- p=5.651e-06
Score (logrank) test = 20.86 on 1 df
- p=4.942e-06
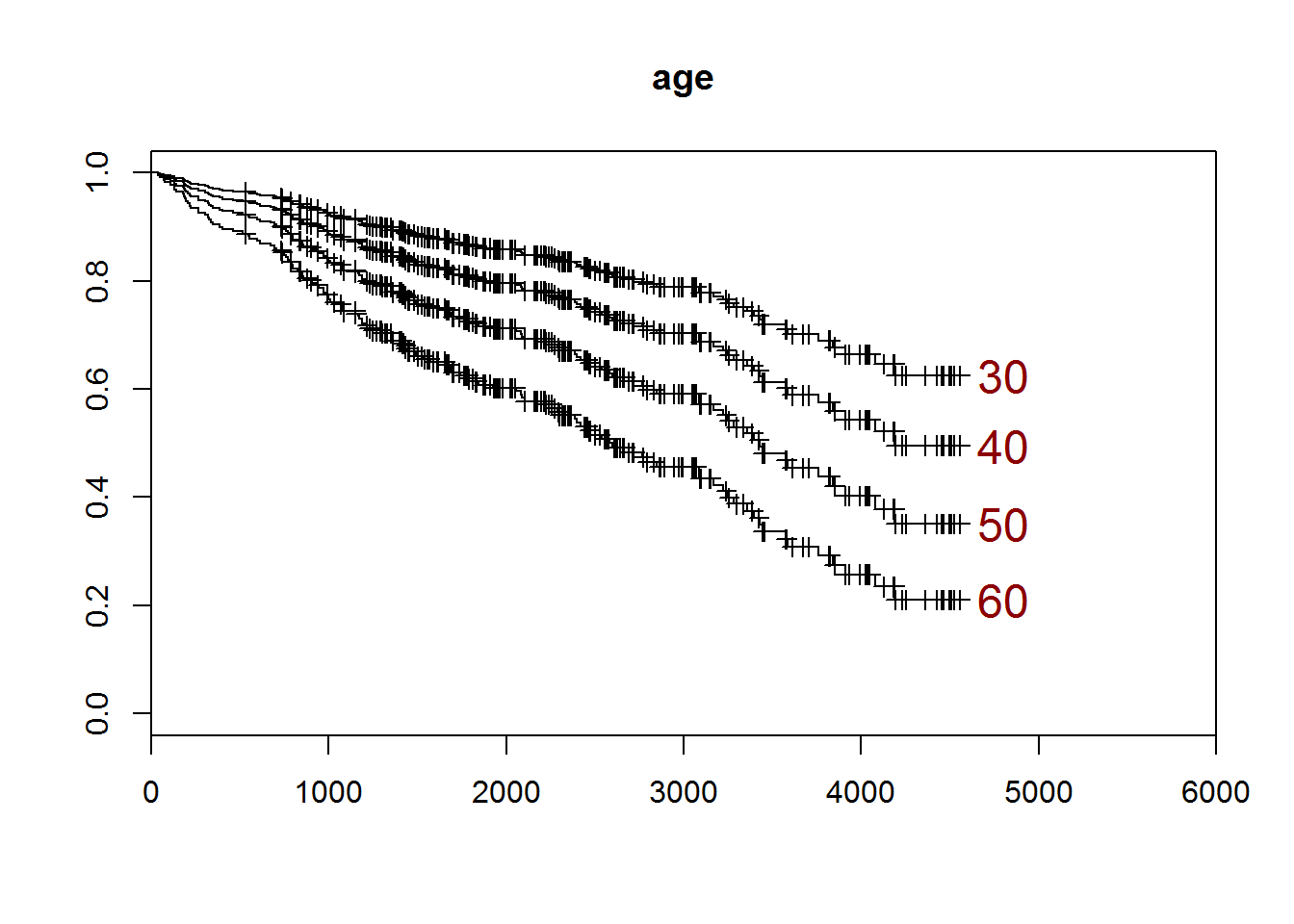
pred.mod2 <- survfit(cox.mod2,newdata=data.frame(age=c(30,40,50,60)))
plot(pred.mod2,xlim=c(0,6000))
title("age")
end.pts <- dim(pred.mod2surv)[1]
end.x <- pred.mod2time[end.pts]+100
end.y <- pred.mod2surv[end.pts,]
end.nm <- c(30,40,50,60)
text(end.x,end.y,end.nm,cex=1.5,col="darkred",adj=0)
You can find an earlier version of this page on my blog.