One of the research projects I am involved with may make use of the HapMap project (www.hapmap.org). This project is an ambitious effort to document the frequency of most Single Nucleotide Polymorphisms (SNPs) in the Human Genome. A SNP is a location on the genome where some subjects show differences in a single nucleotide. The HapMap project looks at four populations:
- CEU, 30 related samples (two parents and an adult offspring) from Utah, USA residents with ancestry from northern and western Europe.
- CHB, 45 unrelated individuals from Han Chinese in Beijing, China.
- JPT, 45 unrelated individuals from Japanese in Tokyo, Japan.
- YRI, 30 related samples (two parents and an adult offspring) from Yoruba in Ibadan, Nigeria.
There is data on all the chromosomes, but I started some analyses using the Y chromosome since it has a very small number of SNPs. In the CEU file, there are 69, in CHB there are 68, in JPT there are 68, and in YRI there are 64. Across all four populations, there are 51 SNPs in common. Some of these SNPs are uninteresting, because 100% of the subjects tested so far had the same nucleotide. Eliminating these SNPs, and you have 15 remaining. Here is the raw data on these 15 SNPs.
rs2534636 2302306 ceu C/C C/T T/T 27 01 28 rs2534636 2302306 chb C/C C/T T/T 20 0 0 20 rs2534636 2302306 jpt C/C C/T T/T 20 0 0 20 rs2534636 2302306 yri C/C C/T T/T 30 0 0 30 97 0 1 98 rs9786608 2309073 ceu C/C C/T T/T 27 0 0 27 rs9786608 2309073 chb C/C C/T T/T 21 0 0 21 rs9786608 2309073 jpt C/C C/T T/T 19 0 0 19 rs9786608 2309073 yri C/C C/T T/T 0 0 28 28 67 0 28 95 rs3899 6994490 ceu G/G G/T T/T 30 00 30 rs3899 6994490 chb G/G G/T T/T 22 00 22 rs3899 6994490 jpt G/G G/T T/T 20 02 22 rs3899 6994490 yri G/G G/T T/T 30 00 30 102 0 2 104 rs9786896 8322494 ceu A/A A/G G/G 0 21 9 30 rs9786896 8322494 chb A/A A/G G/G 0 0 17 17 rs9786896 8322494 jpt A/A A/G G/G 0 0 19 19 rs9786896 8322494 yri A/A A/G G/G 0 0 30 30 0 21 75 96 rs9785941 13105848 ceu A/A A/G G/G 30 0 0 30 rs9785941 13105848 chb A/A A/G G/G 18 0 0 18 rs9785941 13105848 jpt A/A A/G G/G 22 0 0 22 rs9785941 13105848 yri A/A A/G G/G 2 0 28 30 72 0 28 100 rs2032597 13856457 ceu A/A A/C C/C 21 0 8 29 rs2032597 13856457 chb A/A A/C C/C 21 0 0 21 rs2032597 13856457 jpt A/A A/C C/C 22 0 0 22 rs2032597 13856457 yri A/A A/C C/C 29 0 0 29 93 0 8 101 rs2032605 13933534 ceu C/C C/T T/T 28 0 0 28 rs2032605 13933534 chb C/C C/T T/T 20 0 1 21 rs2032605 13933534 jpt C/C C/T T/T 21 0 0 21 rs2032605 13933534 yri C/C C/T T/T 30 0 0 30 99 0 1 100 rs2032590 14028278 ceu T/T G/T G/G 29 0 0 29 rs2032590 14028278 chb T/T G/T G/G 20 0 0 20 rs2032590 14028278 jpt T/T G/T G/G 22 0 0 22 rs2032590 14028278 yri T/T G/T G/G 17 0 9 26 88 0 9 97 rs2032624 14035089 ceu A/A A/C C/C 9 0 21 30 rs2032624 14035089 chb A/A A/C C/C 22 0 0 22 rs2032624 14035089 jpt A/A A/C C/C 22 0 0 22 rs2032624 14035089 yri A/A A/C C/C 30 0 0 30 83 0 21 104 rs2032658 14590648 ceu G/G A/G A/A 21 0 9 30 rs2032658 14590648 chb G/G A/G A/A 0 0 18 18 rs2032658 14590648 jpt G/G A/G A/A 0 0 20 20 rs2032658 14590648 yri G/G A/G A/A 0 0 30 30 21 0 77 98 rs3848982 20612392 ceu G/G A/G A/A 29 0 0 29 rs3848982 20612392 chb G/G A/G A/A 21 0 0 21 rs3848982 20612392 jpt G/G A/G A/A 12 0 9 21 rs3848982 20612392 yri G/G A/G A/A 0 0 30 30 62 0 39 101 rs9306841 20674182 ceu C/C C/G G/G 29 0 0 29 rs9306841 20674182 chb C/C C/G G/G 20 0 0 20 rs9306841 20674182 jpt C/C C/G G/G 18 0 0 18 rs9306841 20674182 yri C/C C/G G/G 0 0 30 30 67 0 30 97 rs2032612 20761675 ceu C/C C/T T/T 30 0 0 30 rs2032612 20761675 chb C/C C/T T/T 20 0 0 20 rs2032612 20761675 jpt C/C C/T T/T 16 0 2 18 rs2032612 20761675 yri C/C C/T T/T 30 0 0 30 96 0 2 98 rs2032635 20784951 ceu A/A A/G G/G 19 0 9 28 rs2032635 20784951 chb A/A A/G G/G 0 0 22 22 rs2032635 20784951 jpt A/A A/G G/G 0 0 22 22 rs2032635 20784951 yri A/A A/G G/G 0 0 30 30 19 0 83 102 rs2032652 20812497 ceu T/T C/T C/C 30 0 0 30 rs2032652 20812497 chb T/T C/T C/C 20 0 2 22 rs2032652 20812497 jpt T/T C/T C/C 10 0 12 22 rs2032652 20812497 yri T/T C/T C/C 0 0 30 30 60 0 44 104
The first entry is the identification number for the SNP, and the second
entry gives you the physical location of the SNP in base pairs. Notice
that even though the Y chromosome is very small, there are still tens of
millions of base pairs. The third entry identifies the group. The
fourth, fifth, and sixth entries give the genotype for the SNP. The
HapMap project uses two letters for consistency with the rest of the
chromosome, but Y (and X in males) occurs by itself, so it is impossible
to have a heterogenous SNP. The fourth SNP (rs9786896) has an obvious
typo, since it includes counts for a heterogenous SNP.
An interesting question is whether the value of a base at particular SNPs provides any useful clues about whether a person might belong to CEU, CHB, JPT, or YRI. For a particular SNP to be helpful, it must show a range of variation. The first SNP (rs2534636) is T for 97 out of 98 people, and is unlikely to produce any valuable clues. In contrast, the second SNP (rs9786608) is C for 67 cases and T for 28 cases.
You can use a measure of uncertainty (sometimes called the entropy measure) to quantify the variation in SNP probabilities. The formula for uncertainty is
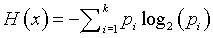
where pi represent the probabilities of the two base pairs (or the probabilities of the three possible allele combinations for paired genes). The uncertainty measures for the 15 SNPs are:
rs# position H(Y) rs2534636 2302306 0.08 rs9786608 2309073 0.87 rs3899 6994490 0.14 rs9786896 8322494 0.76 rs9785941 13105848 0.86 rs2032597 13856457 0.40 rs2032605 13933534 0.08 rs2032590 14028278 0.45 rs2032624 14035089 0.73 rs2032658 14590648 0.75 rs3848982 20612392 0.96 rs9306841 20674182 0.89 rs2032612 20761675 0.14 rs2032635 20784951 0.69 rs2032652 20812497 0.98
Notice that the uncertainty is largest for SNPs with the closest to 50-50 splits. The last SNP (rs2032652), for example, has 60 Ts and 44 Cs and has an uncertainty of 0.98.
So showing a range of variation, or a large value of uncertainty is helpful for distinguishing among the groups, but it is also important to show that this variation or uncertainty is dependent on the groups themselves. The concept of conditional uncertainty is helpful for quantifying this. The conditional uncertainty is computed in a fashion similar to the uncertainty measure described above, but you substitute conditional probabilities in the formula:
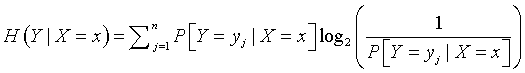
You can compute conditional uncertainty for each group. The results are presented below:
rs# position group H(Y|x) rs2534636 2302306 ceu 0.22 rs2534636 2302306 chb 0.00 rs2534636 2302306 jpt 0.00 rs2534636 2302306 yri 0.00 rs9786608 2309073 ceu 0.00 rs9786608 2309073 chb 0.00 rs9786608 2309073 jpt 0.00 rs9786608 2309073 yri 0.00 rs3899 6994490 ceu 0.00 rs3899 6994490 chb 0.00 rs3899 6994490 jpt 0.44 rs3899 6994490 yri 0.00 rs9786896 8322494 ceu 0.88 rs9786896 8322494 chb 0.00 rs9786896 8322494 jpt 0.00 rs9786896 8322494 yri 0.00 rs9785941 13105848 ceu 0.00 rs9785941 13105848 chb 0.00 rs9785941 13105848 jpt 0.00 rs9785941 13105848 yri 0.35 rs2032597 13856457 ceu 0.85 rs2032597 13856457 chb 0.00 rs2032597 13856457 jpt 0.00 rs2032597 13856457 yri 0.00 rs2032605 13933534 ceu 0.00 rs2032605 13933534 chb 0.28 rs2032605 13933534 jpt 0.00 rs2032605 13933534 yri 0.00 rs2032590 14028278 ceu 0.00 rs2032590 14028278 chb 0.00 rs2032590 14028278 jpt 0.00 rs2032590 14028278 yri 0.93 rs2032624 14035089 ceu 0.88 rs2032624 14035089 chb 0.00 rs2032624 14035089 jpt 0.00 rs2032624 14035089 yri 0.00 rs2032658 14590648 ceu 0.88 rs2032658 14590648 chb 0.00 rs2032658 14590648 jpt 0.00 rs2032658 14590648 yri 0.00 rs3848982 20612392 ceu 0.00 rs3848982 20612392 chb 0.00 rs3848982 20612392 jpt 0.99 rs3848982 20612392 yri 0.00 rs9306841 20674182 ceu 0.00 rs9306841 20674182 chb 0.00 rs9306841 20674182 jpt 0.00 rs9306841 20674182 yri 0.00 rs2032612 20761675 ceu 0.00 rs2032612 20761675 chb 0.00 rs2032612 20761675 jpt 0.50 rs2032612 20761675 yri 0.00 rs2032635 20784951 ceu 0.91 rs2032635 20784951 chb 0.00 rs2032635 20784951 jpt 0.00 rs2032635 20784951 yri 0.00 rs2032652 20812497 ceu 0.00 rs2032652 20812497 chb 0.44 rs2032652 20812497 jpt 0.99 rs2032652 20812497 yri 0.00
For some SNPs, the conditional uncertainties are larger than the (unconditional) uncertainty for one group and smaller for the other three groups. For other SNPs, the conditional uncertainties are all smaller than the unconditional uncertainty. You can aggregate the conditional uncertainties together using the formula:
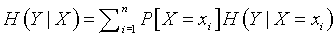
This is just a weighted average of the conditional uncertainties. The difference between the conditional uncertainty and the unconditional uncertainty is called the mutual information.
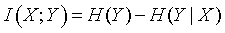
This definition is quite intuitive. The mutual information represents the decrease in uncertainty in Y when we know the value of X. This definition of mutual information is symmetric, so if you define
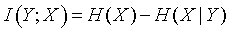
then you will get the exact same answer
cc
rs# position H(Y) H(Y|X) I(X;Y) rs2534636 2302306 0.08 0.06 0.02 rs9786608 2309073 0.87 0.00 0.87 rs3899 6994490 0.14 0.09 0.04 rs9786896 8322494 0.76 0.28 0.48 rs9785941 13105848 0.86 0.11 0.75 rs2032597 13856457 0.40 0.24 0.16 rs2032605 13933534 0.08 0.06 0.02 rs2032590 14028278 0.45 0.25 0.20 rs2032624 14035089 0.73 0.25 0.47 rs2032658 14590648 0.75 0.27 0.48 rs3848982 20612392 0.96 0.20 0.76 rs9306841 20674182 0.89 0.00 0.89 rs2032612 20761675 0.14 0.09 0.05 rs2032635 20784951 0.69 0.25 0.44 rs2032652 20812497 0.98 0.30 0.68
Things left to do: Add commentary about SNPs with high information.
There is a lot of discussion about linkage disequilibrium, and these measures of joint and conditional uncertainty may provide some useful ways to assess the degree of disequilibrium. Here are a few references about disequilibrium that relate to the HapMap project:
- Finding genes underlying risk of complex disease by linkage disequilibrium mapping. Clark AG. Curr Opin Genet Dev 2003: 13(3); 296-302. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12787793&dopt=Abstract) [PDF]](http://genetics.med.harvard.edu/genetics201/Clark.pdf)
- Efficient computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies. Dudbridge F, Koeleman BP. Am J Hum Genet 2004: 75(3); 424-35. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15266393&dopt=Abstract) [Abstract]](http://www.journals.uchicago.edu/cgi-bin/resolve?AJHG41328ABS) [Full text]](http://www.journals.uchicago.edu/AJHG/journal/issues/v75n3/41328/41328.html) [PDF]](http://www.journals.uchicago.edu/AJHG/journal/issues/v75n3/41328/41328.web.pdf)
- Human haplotype block sizes are negatively correlated with recombination rates. Greenwood TA, Rana BK, Schork NJ. Genome Res 2004: 14(7); 1358-61. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15231751&dopt=Abstract) [Abstract]](http://www.genome.org/cgi/content/abstract/14/7/1358) [Full text]](http://www.genome.org/cgi/content/full/14/7/1358) [PDF]](http://www.genome.org/cgi/reprint/14/7/1358.pdf)
- Linkage disequilibrium maps and association mapping. Morton NE. J Clin Invest 2005: 115(6); 1425-30. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15931377&dopt=Abstract) [Abstract]](http://www.jci.org/cgi/content/abstract/115/6/1425) [Full text]](http://www.jci.org/cgi/content/full/115/6/1425) [PDF]](http://www.jci.org/cgi/reprint/115/6/1425.pdf)
- A map of the human genome in linkage disequilibrium units. Tapper W, Collins A, Gibson J, Maniatis N, Ennis S, Morton NE. Proc Natl Acad Sci U S A 2005: 102(33); 11835-9. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16091463&dopt=Abstract) [Full text]](http://www.blackwell-synergy.com/links/doi/10.1046/j.1469-1809.2003.00050.x/full)
- A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Evans DM, Cardon LR. Am J Hum Genet 2005: 76(4); 681-7. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15719321&dopt=Abstract) [Full text]](http://www.journals.uchicago.edu/AJHG/journal/issues/v76n4/41887/41887.html) [PDF]](http://www.journals.uchicago.edu/AJHG/journal/issues/v76n4/41887/41887.web.pdf)
Additional reference (added January 3, 2006)
- Genomic scans for selective sweeps using SNP data. Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genome Res 2005: 15(11); 1566-75. [Medline]](http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16251466&dopt=Abstract) [Abstract]](http://www.genome.org/cgi/content/abstract/15/11/1566) [Full text]](http://www.genome.org/cgi/content/full/15/11/1566) [PDF]](http://www.genome.org/cgi/reprint/15/11/1566.pdf)
You can find an earlier version of this page on my original website.